WISpR: Mapping the Cellular Puzzle of Life
A machine learning approach reveals hidden patterns in spatial transcriptomics data
Nuray Sogunmez Erdogan and Deniz Eroglu
Kadir Has University, Istanbul
Published in PLOS Computational Biology
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1013169
Every cell in our body plays a role — but finding out where each one belongs is like solving a giant puzzle. To understand how our organs grow, how diseases begin, or how treatments might work, we need to know not just which cells are present, but how they are organized together.
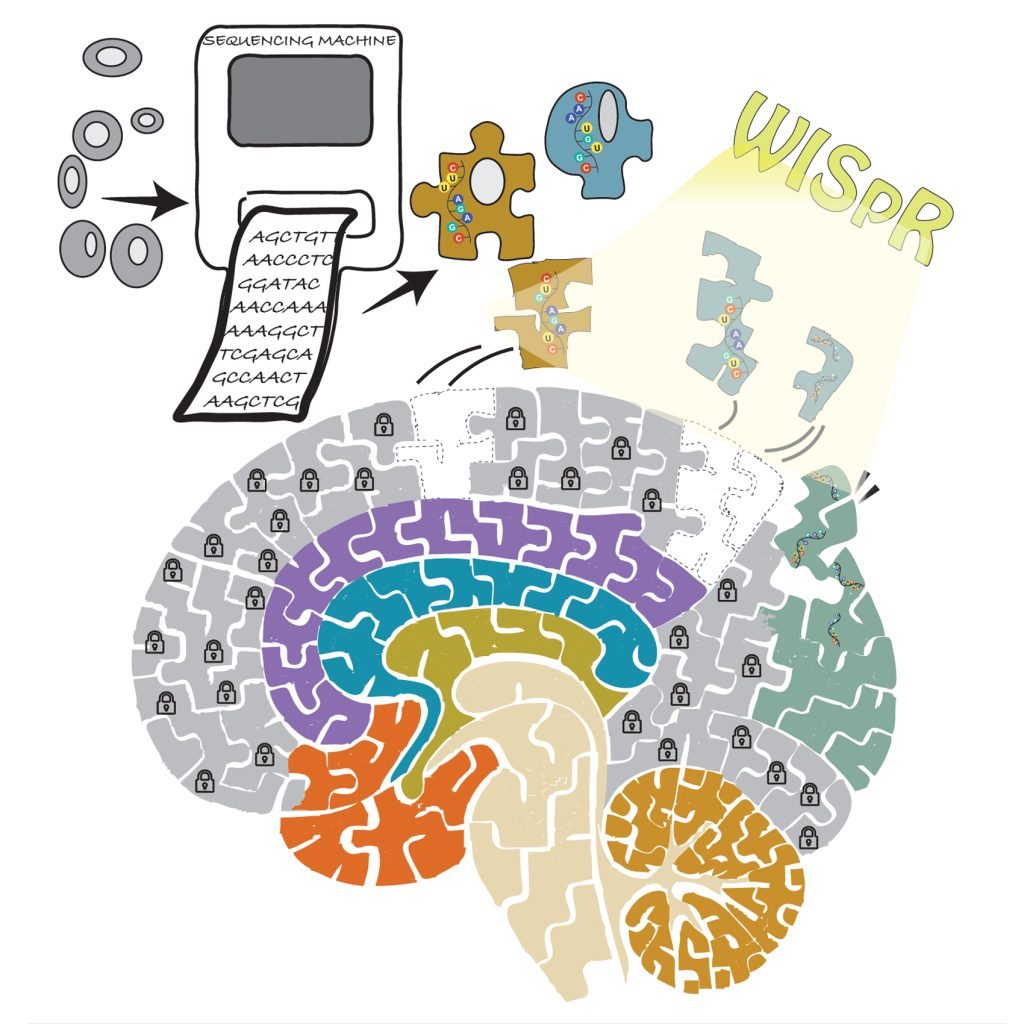
| Figure 1: WISpR bridges cell-level resolution and spatial organization by decoding the biolog-ical puzzle—one cell at a time. |
Think of attempting to finish a 1,000-piece jigsaw puzzle with- out a picture to guide you. Some pieces are easily recognizable, such as the borders or those with bright designs, while others are unclear, puzzling, or appear not to fit in at all. This is similar to the experience of researchers who try to understand the arrangement of various cell types within our tissues. Just as a single lost or incorrectly placed puzzle piece can distort the overall picture, an inaccurate interpretation of cell organization can hinder our understanding of how diseases form and progress.
To tackle this challenge, researchers Nuray Sogunmez Erdogan and Deniz Eroglu at Kadir Has University have introduced WISpR (Weight-Induced Sparse Regression), a novel machine learning method that interprets noisy biological data and con- structs a more precise and clear representation of how cells are structured within our tissues.
The Puzzle of the Human Body
Our bodies consist of trillions of cells that originate from a single fertilized egg, carefully arranged into tissues and organs with remarkable precision. Even within a single tissue, numerous unique cell types are strategically distributed, communicating via chemical signals, constructing structures, and reacting to external stimuli by reporting themselves using their own language, gene expression.
These cells are aware of their functions and designated locations, yet it remains a mystery for researchers to replicate this intricate organization. Identifying how these cells are located and how their arrangement alters in conditions such as cancer, heart abnormalities, or neurodegenerative diseases is vital. However, this task is quite challenging.
Two recent technologies have emerged to address this challenge by translating the complex language of cellular gene expression into terms humans can understand:
- Spatial transcriptomics offers a layout showing where genes are active in slices of tissue; however, each area might contain various cell types, complicating the identification of individual roles.
- Single-cell RNA sequencing (scRNA-Seq) reveals what each specific cell is producing, but it detaches them from their original environment, similar to examining puzzle pieces without having the complete picture.
So, how do we integrate these components?
Why this matters?
In medicine and biology, location is everything. Knowing that a particular immune cell is present in a tumor is not enough. We need to know where it is. Is it close to blood vessels? Neighboring a cancer stem cell? Or are they buried deep in necrotic tissue?
Producing high-resolution, interpretable maps of tissue composition opens the door to better disease diagnostics by identifying microenvironmental niches, understanding development by mapping dynamic transitions in fetal tissues, and tracking disease progression by visualizing cellular changes over time. Applications range from basic research to clinical translation.
What is overseen till now?
Researchers have gone back to computational methods called “deconvolution tools” to link these two types of data. Using data from single-cell RNA sequencing as a guide, these instruments attempt to identify the types of cells present at each given location. However, many modern techniques overpredict the range of cell types found in every position. They view every conceivable cell type as a possibility, even if the signal is faint or insignificant. This can produce biologically implausible results when rare or unusual cell types are discovered in locations where they should not be. Other problems that need to be solved include sensitivity to variances between datasets (variances across laboratories or platforms), ignoring the specific distribution of cell types within tissues (since not every cell type is found everywhere), and challenges related to noisy or mismatched data, which often occurs when public references are used. As a result, many deconvolution programs often produce cell maps that confuse rather than provide insightful and accurate information.
WISpR: A Smarter, Leaner Way to Deconvolute Tissues
WISpR is an innovative deconvolution technique created by a molecular biologist and a mathematician to address these issues. WISpR offers a mathematically sound and biologically informed approach to choose the most probable and informative cell types at each spatial point. WISpR incorporates a penalty that encourages sparsity, which diminishes background noise and signals that are not strongly supported, concentrating only on biologically relevant inputs. Even when the reference and target datasets originate from different organisms, times, or research projects, WISpR maintains strong performance, due to its robustness against batch variations and disparities. The resulting outputs from WISpR are clear and reliable maps that accurately represent cell compositions, facilitating the extraction of biological insights. The tool was thoroughly tested with both simulated data and actual datasets, including developing human heart, mouse brain, and breast cancer samples. In all cases, it outperformed leading tools in terms of accuracy, sparsity, and biological relevance.
Looking Ahead: WISpR in Action
As tissue atlases grow in size and complexity and as spatial technologies become more accessible, tools such as WISpR will be essential to translate raw data into meaningful knowledge. In the future, WISpR could help discover rare cell populations hiding in disease hotspots to predict patient outcomes by mapping immune infiltration in tumors, guide regenerative therapies by identifying where to deliver stem cells, and clarify how cells interact in development, repair, or degeneration. In short, WISpR does not just see the pieces; it knows where they belong. And when we know how the pieces fit together, we can begin to solve the larger puzzle of life.
“Each cell-type driven by gene expression is a puzzle piece. WISpR helps reveal the full picture.”
